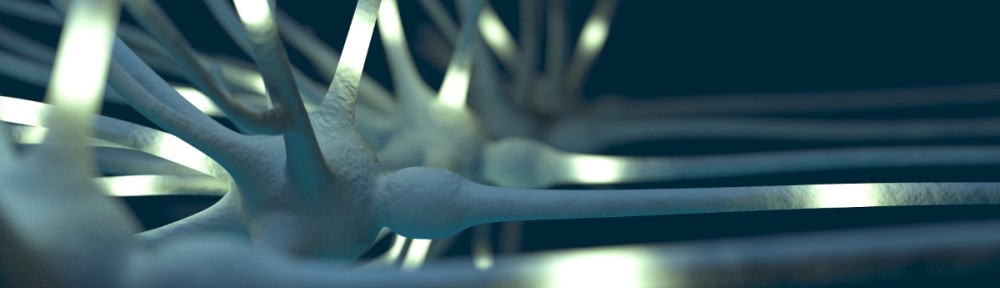
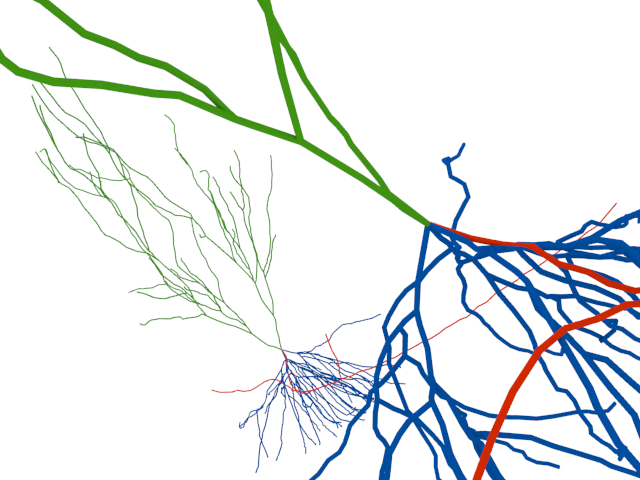
This is another interesting use-case of Blender in neuroscience to study the elements and statistical properties of neural tissue from data obtained with an electron microscope.
Advances in the application of electron microscopy (EM) to serial imaging are opening doors to new ways of analyzing cellular structure. New and improved algorithms and workflows for manual and semiautomated segmentation allow us to observe the spatial arrangement of the smallest cellular features with unprecedented detail in full three-dimensions. From larger samples, higher complexity models can be generated; however, they pose new challenges to data management and analysis. Here we review some currently available solutions and present our approach in detail. We use the fully immersive virtual reality (VR) environment CAVE (cave automatic virtual environment), a room in which we are able to project a cellular reconstruction and visualize in 3D, to step into a world created with Blender, a free, fully customizable 3D modeling software with NeuroMorph plug-ins for visualization and analysis of EM preparations of brain tissue. Our workflow allows for full and fast reconstructions of volumes of brain neuropil using ilastik, a software tool for semiautomated segmentation of EM stacks. With this visualization environment, we can walk into the model containing neuronal and astrocytic processes to study the spatial distribution of glycogen granules, a major energy source that is selectively stored in astrocytes. The use of CAVE was key to the observation of a nonrandom distribution of glycogen, and led us to develop tools to quantitatively analyze glycogen clustering and proximity to other subcellular features. J. Comp. Neurol., 2015. © 2015 Wiley Periodicals, Inc.