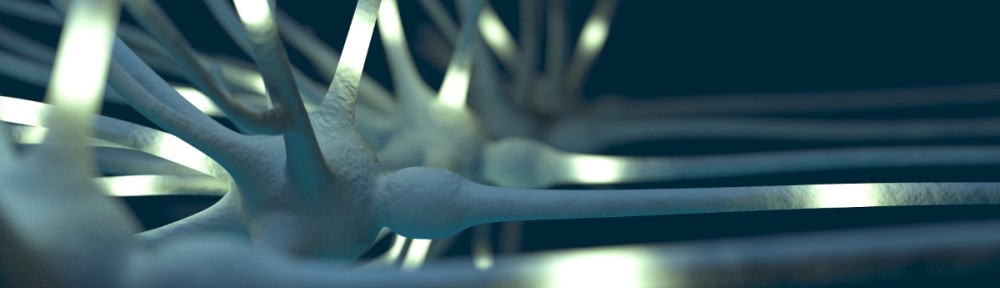
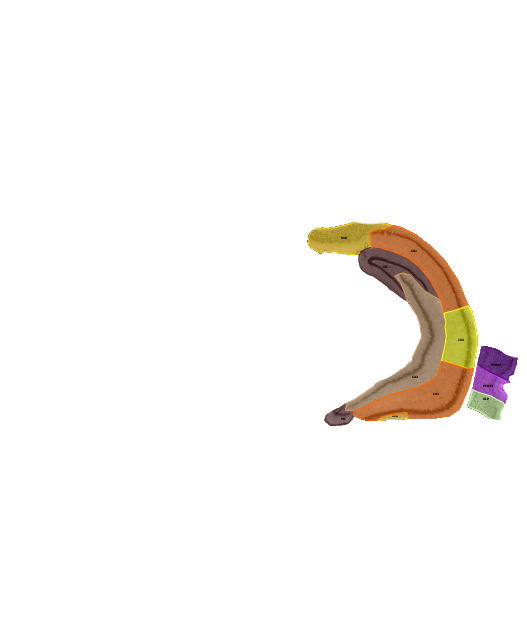I think the hippocampal formation is one of the hardest regions to understand with regard to its overall shape and connection patterns. In the rodent it crosses all three axes in the brain and the orientation of its convoluted layers changes across its septal-temporal axis. Therefore, I am happy to share with you a video that I produced recently, in which I show how the hippocampal formation looks like in 3d. For me it was also a good exercise to produce a video for Youtube, as I am going to produce more in the next weeks.
Author Archives: admin
Blender Addon: Map via UV
Here is a little operator that I wrote for Blender. This operator is basically a side-result of a larger project I am working on but I thought it is worth sharing this as it might be useful for others as well.
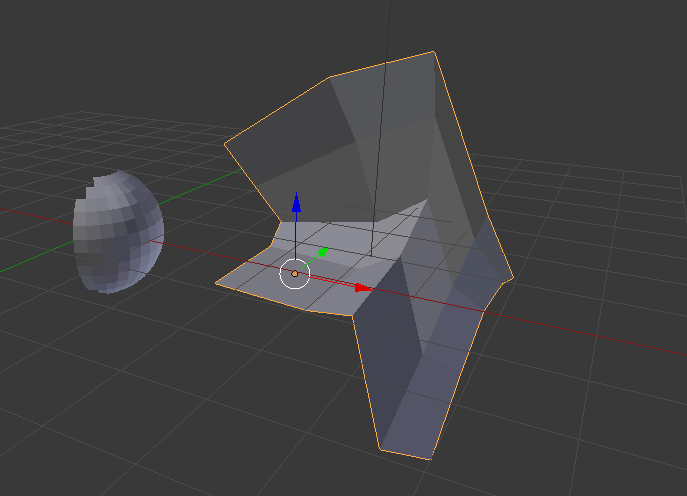
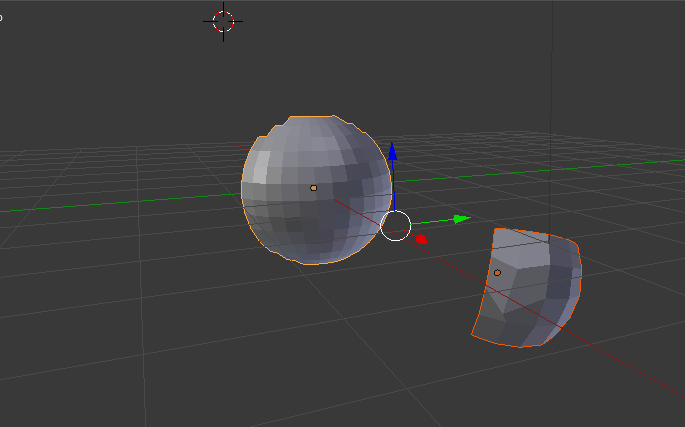
The aim is to bring a given mesh-surface into another shape. For example, in the picture below, I would like to transform the right mesh (the origin) into the form of the left mesh (the target) while its internal topology should be maintained.
I found that the easiest way to achieve this is, to unwrap both objects and simply define in UV-space how the right mesh should cover the left mesh.
The only important point here is that any point in the UV-map of the origin mesh must be in the area of the UV-map of the target-mesh. Then, hit “map via UV” and you got
The addon and additional information as well as some debugging issues can be found and discussed in this BlenderArtists-Thread.
Jeff Lichtman shows the complexity of the brain
This video is fascinating and frustrating (from a computational modeling point of view) at the same time. Jeff Lichtman shows in this video (above) the complexity of a tiny little fraction of the mouse brain scanned on a nanometer scale using serial electron microscopy. At 16:00 and the following minutes he shows the relation between the nanometer scale, the complexity that can be observed on this level and the large scale view on the brain. Absolutely impressive.
Interesting Articles (1)
Functional connectivity of the entorhinal-hippocampal space circuit by Sheng-Jia Zhang and colleagues from the Kavli Institute for Systems Neuroscience in Trondheim.
They used light-evoked discharges of grid- and other cells to determine whether these cells have direct hippocampal projections. What I find interesting about this study is that principle cells in MEC need between <5-30ms to fire, depending on the current they receive.
New and Distinct Hippocampal Place Codes Are Generated in a New Environment during Septal Inactivation by Mark Brandon and colleagues from the Center for Functional Connectomics in Seoul.
In this paper they show that septal inactivation leads to reduced theta power and grid cell activity. In a novel environment place fields emerged which suggests (according to their interpretation) that neurons can generate place fields without grid cell activity.
Works related to Novelty Search
Novelty Search has been proposed as alternative to objective driven search in evolutionary runs. It has been found that in some problem domains, Novelty Search performs better than an objective driven search in finding a spot in a large but deceptive solution space. This list links the general idea behind Novelty Search to works from other disciplines which go in a similar direction.
Read VTK files from the Allen Brain Atlas in Blender
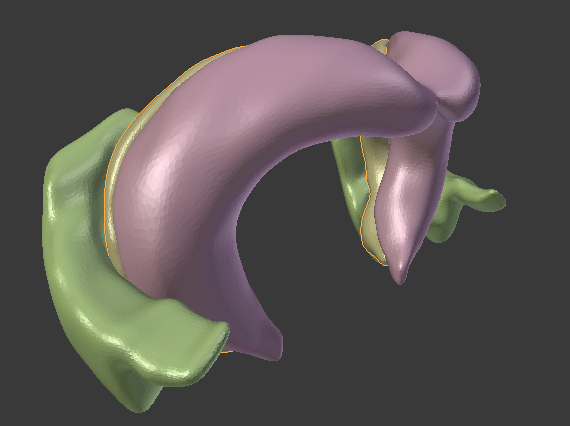
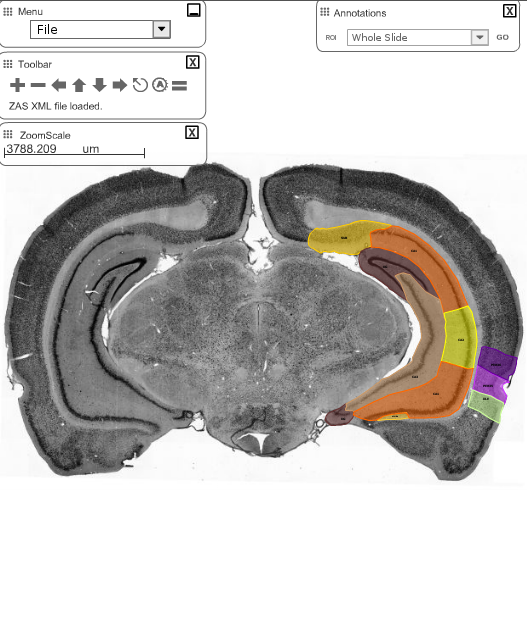
The Allen Brain Atlas provides most of their tracked brain data as 3d-files in the VTK-format. I found it difficult to find a converter for this format that would give me e.g. an obj-file. I came across the Graph Toolbox for Matlab which can read vtk-files and write obj-files. However, their importer did not work for that particular version of the vtk-format. Here is read_Allen_vtk.m, an m-file that I derived from read_vtk.m, which is able to extract all vertices and face-data from the vtk-files. You can then save them using write_obj() and open them in the 3d-tool of your choice. In the upper image you see the hippocampus and entorhinal cortex of the mouse in Blender.
Masking with Matlab
Recently, I had a bunch of images for which I wanted to make all grey pixels transparent, in order to just see the color coded regions.
This was one of the situations, in which I was wondering whether I should do it manually (for 37 images) with Gimp, or write a script that can do the job for me. I was happy, that I decided to write a script, because it turned out that it just took me one minute to figure out the commands in Matlab and an additional minute to write the script. At the end I saved a lot of time and obtained images with an even more accurate mask than those images that I produced with Gimp. Here is the script for this kind of job:
function maskImage(filename)
a = imread(filename);
c = ((a(:,:,1) == a(:,:,2)).* (a(:,:,2) == a(:,:,3))) == 0;
imwrite(a, filename, 'png', 'Alpha', double(c));
Molecular Movies
Molecular Movies is a collection of animations of molecular and cell reactions. I stumbled upon this side when I was looking for 3d depictions of the hippocampus. A very nice animation by Drew Berry can be found here and here on Youtube.
Night photography
I recently had the opportunity to go out for a night photography session and became a fan of light painting.
Blender in Science – Part 2
More Blender-related science staff. The extension Add 3D Function Surface and BlenderSciViz allow to plot mathematical functions and external data in Blender.

Mark Meyer uses Blender to explore and explain colorspaces.
And “CellBlender is an addon for Blender 2.6x that allows users to create computational cell biology models for use in MCell.”